Expanded Research Overview
Our research addresses questions related to the responses of microbial populations, communities, and ecosystems to changes in the resource environment. Specifically, we ask questions related to how the supply and diversity of resources alters the composition and function of communities, and how the traits and species interactions link structure and function. To answer these questions, we use a mixture of field, lab, bioinformatics, and theoretical approaches. Using these diverse approaches our research bridges microbiology and community and ecosystem ecology.
Microbial Metabolism
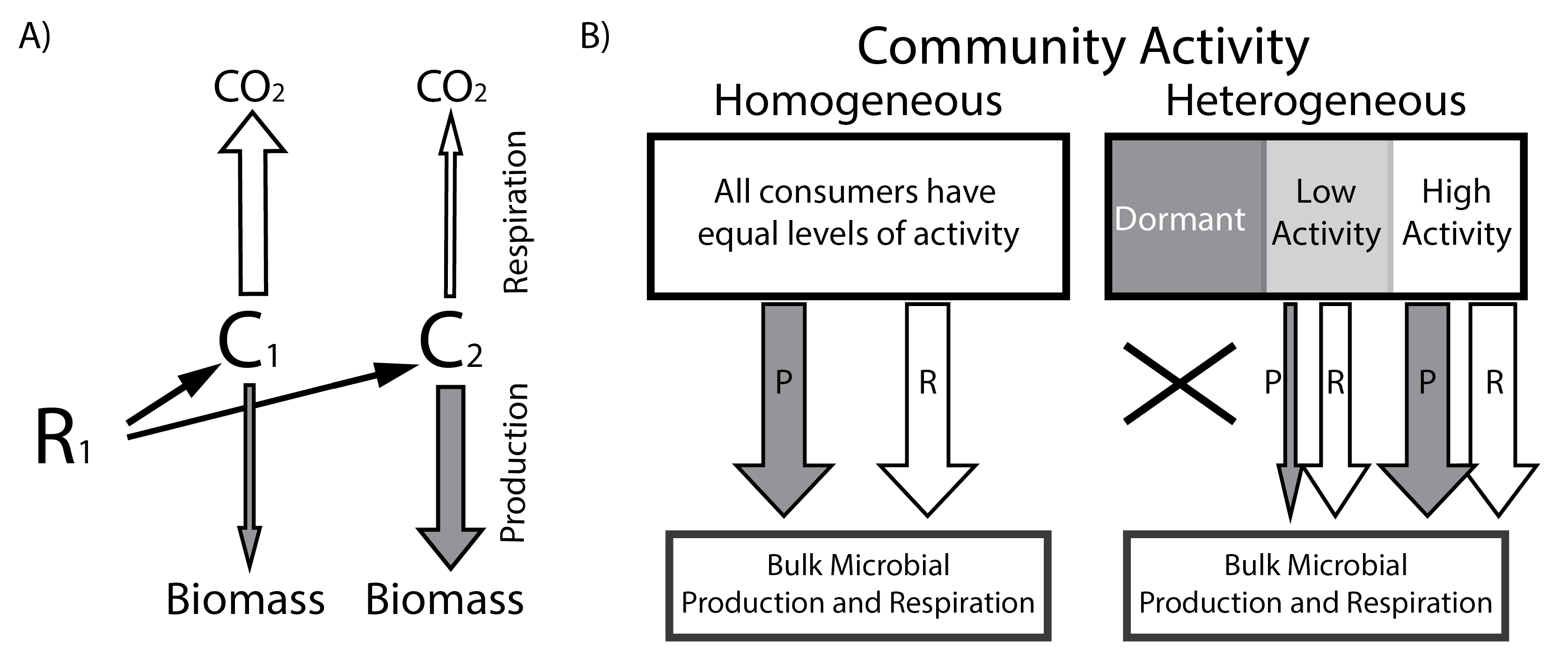
Microbial metabolic rates link the composition of microbial communities to aquatic carbon dynamics. Based on previous studies, there are a number of abiotic and biotic factors which may control microbial metabolism, including physical properties such as pH and hydrological flow and biotic factors such as species interactions in the microbial community. However, there have been no large-scale studies conducted to understand these connections. Because microbial metabolism is the missing gap linking the structure and function of microbial communities, understanding drivers of microbial metabolism will ultimately help constrain predictive models.
Resource Heterogeneity

While often considered a homologous pool, many resources exist as heterogeneous mixtures of multiple chemical forms.
Microbial responses to resource heterogeneity
Using experimental mesocosms, I have explored the resource heterogeneity hypothesis using phosphorus resources. My research shows that different phosphorus resources (e.g., phosphate, phytic acid) alter the diversity and function of aquatic bacterial communities in predictable ways (Muscarella et al. 2014). Based on these findings, I proposed that the myriad of chemical forms representing any resource pool can be grouped into functional categories. With the phosphorus resources, a grouping based on a biomolecules model best predicts the observed data.
I have also applied the resource heterogeneity framework to bulk organic matter in aquatic ecosystems. Organic matter is used by heterotrophic microorganism to meet their metabolic demands. Using high-resolution mass spectrometry, I cataloged the diversity of organic matter in lakes, and demonstrated that organic matter composition can explain 40% of the diversity in heterotrophic microbial communities. I hypothesize that resource heterogeneity and resource acquisition strategy (i.e., generalists versus specialists) explain resource – diversity relationships.
Related Manuscripts:
-
Muscarella ME, Bird KC, Larsen ML, Placella SA, Lennon JT (2014). Phosphorus resource heterogeneity affects the structure and function of microbial food webs. Aquatic Microbial Ecology, 73(3):259-272
-
Muscarella ME, Boot CM, Broeckling C., Lennon JT (2019). Resource diversity structures aquatic bacterial communities. bioRxiv preprint available, DOI: https://doi.org/10.1101/387803
Microbial responses to altered resource inputs
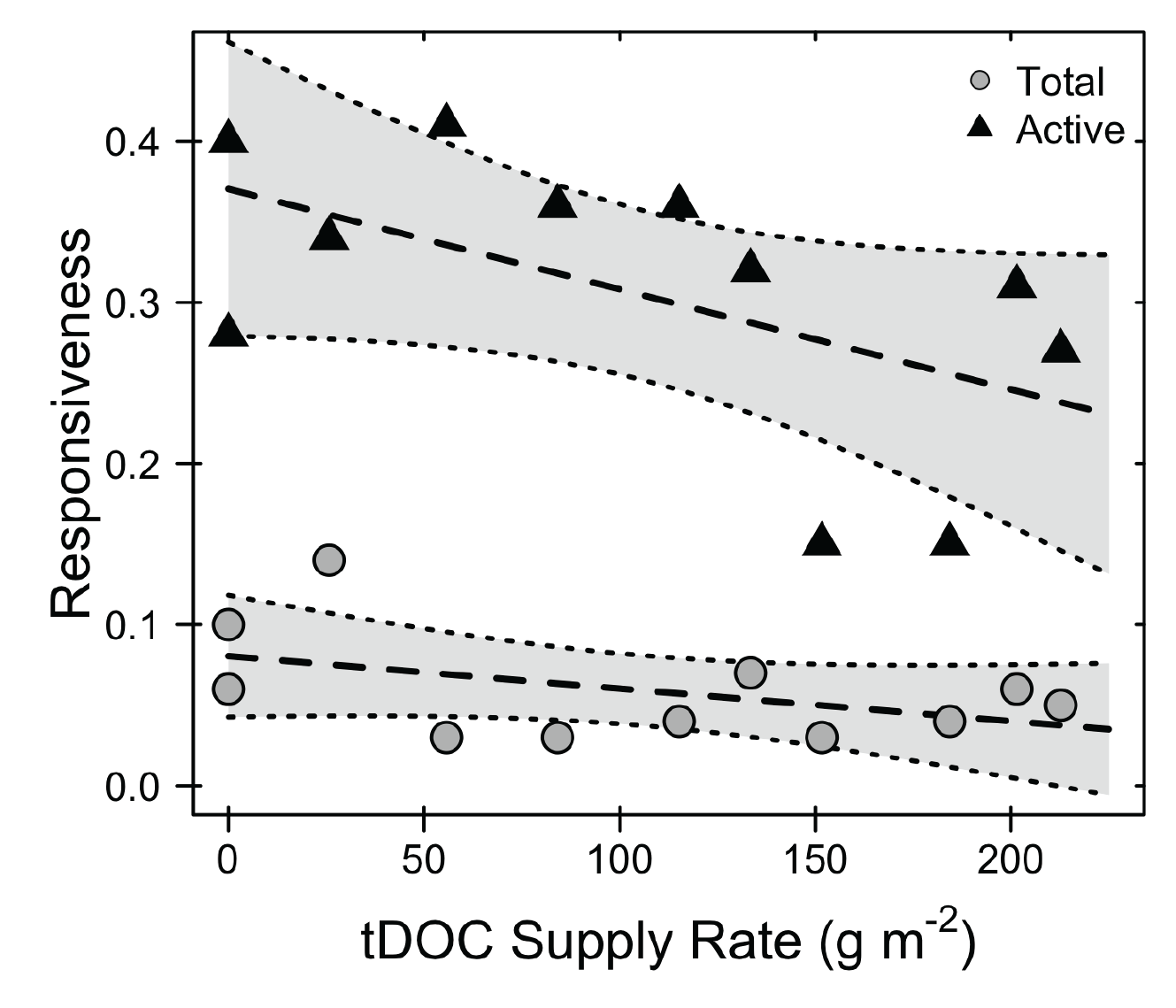
One of the major impacts humans have had on the landscape is changing the inputs of resources. These altered inputs are particularly noticeable in aquatic environments where they are often linked to deteriorating water quality. My research has focused on the altered flow of dissolved organic carbon between terrestrial and aquatic ecosystems. I have explored the impact of tDOC loading on aquatic microbial communities using the experimental ponds at the Kellogg Biological Station. My research shows that tDOC inputs select for specialized microorganisms that use tDOC to meet their metabolic demands. However, based on the dynamics of these specialists, it appears that the community does not rapidly respond to nutrient perturbations and thus can destabilize ecosystem function.
Related Manuscripts:
-
Lennon JT, Hamilton SK, Muscarella ME, AS Grandy, K Wickings, SE Jones (2013). A source of terrestrial organic carbon to investigate the browning of aquatic ecosystems. PLoS ONE 8(10):e75771
-
Muscarella ME, Jones SE, Lennon JT (2016) Species sorting along a subsidy gradient alters bacterial community stability. Ecology, 97(8):2034-2043
Traits and Theory
Microbial Carbon Traits
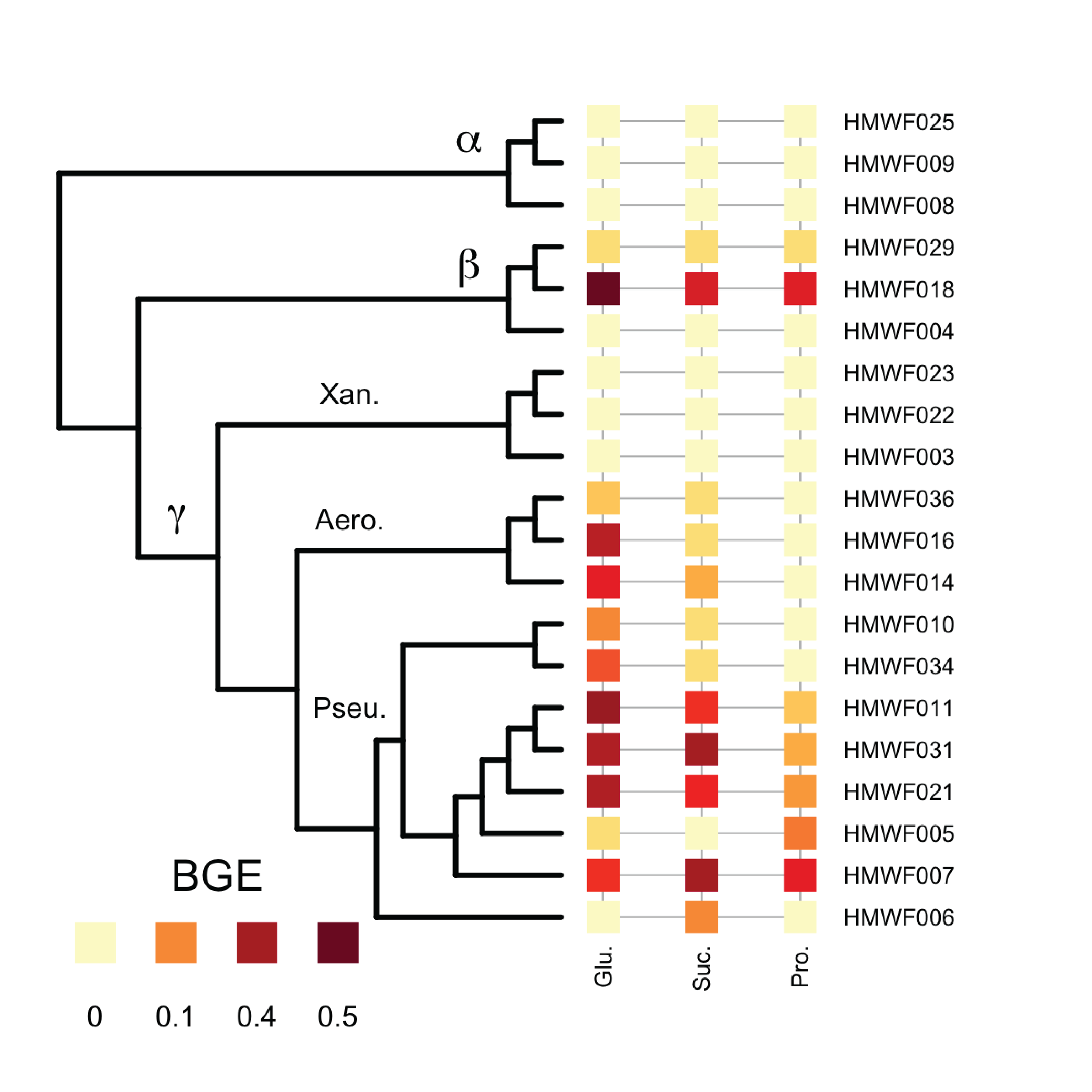
Heterotrophic bacteria in ecosystems determine whether organic carbon is respired and lost as CO_2 or converted into biomass and retained in food webs. Many factors control how bacteria process carbon, but perhaps the most important factor is bacterial growth efficiency (BGE). I measured BGE in a set of bacterial isolates. I use different carbon substrates to represent different types of resources available to heterotrophic bacteria. Overall, I have found that variation in BGE is better explained by consumer identity, and I have also identified physiological trade-offs related to growth strategy. These results suggest that large changes in community composition or available resources may alter BGE in predictive ways.
Related Manuscripts:
- Muscarella ME, Lennon JT (In Review). Trait-based approach to bacterial growth efficiency. bioRxiv preprint available, DOI: https://doi.org/10.1101/427161
Microbial Interactions
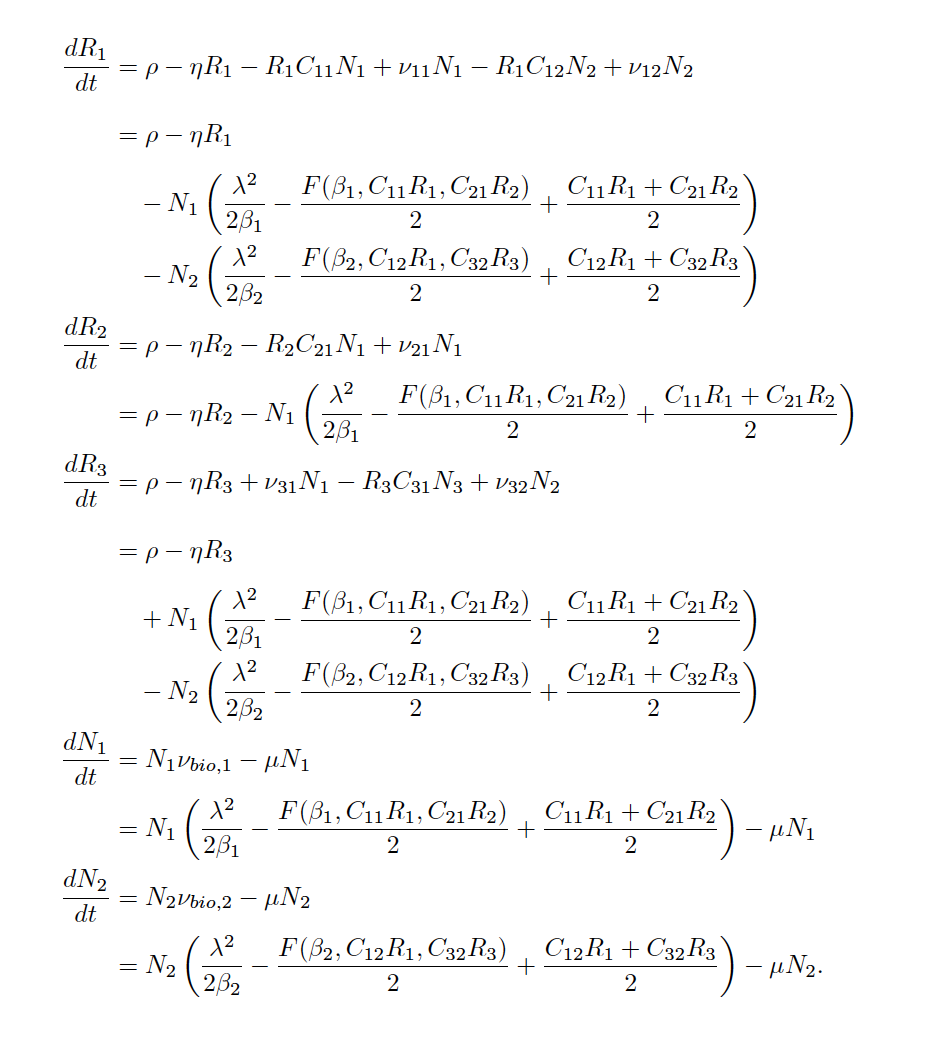
Quantifying the strength, sign, and origin of species interactions, along with their dependence on environmental context, is at the heart of prediction and understanding in ecological communities. We are modeling a coarse-grained version of a species’ true, cellular metabolism to describe resource consumption via uptake and conversion into biomass, energy, and byproducts. Using mathematical models, we have been exploring how secondary metabolite production can support coexistence even when two species compete for a shared resource. Building on these findings, we make the case for incorporating metabolism to update the phenomenology we use to model species interactions.
Related Manuscripts:
- Muscarella ME, O’Dwyer JP (In Review) Species Dynamics and Interactions via Metabolically Informed Consumer-Resource Models
Evolutionary History
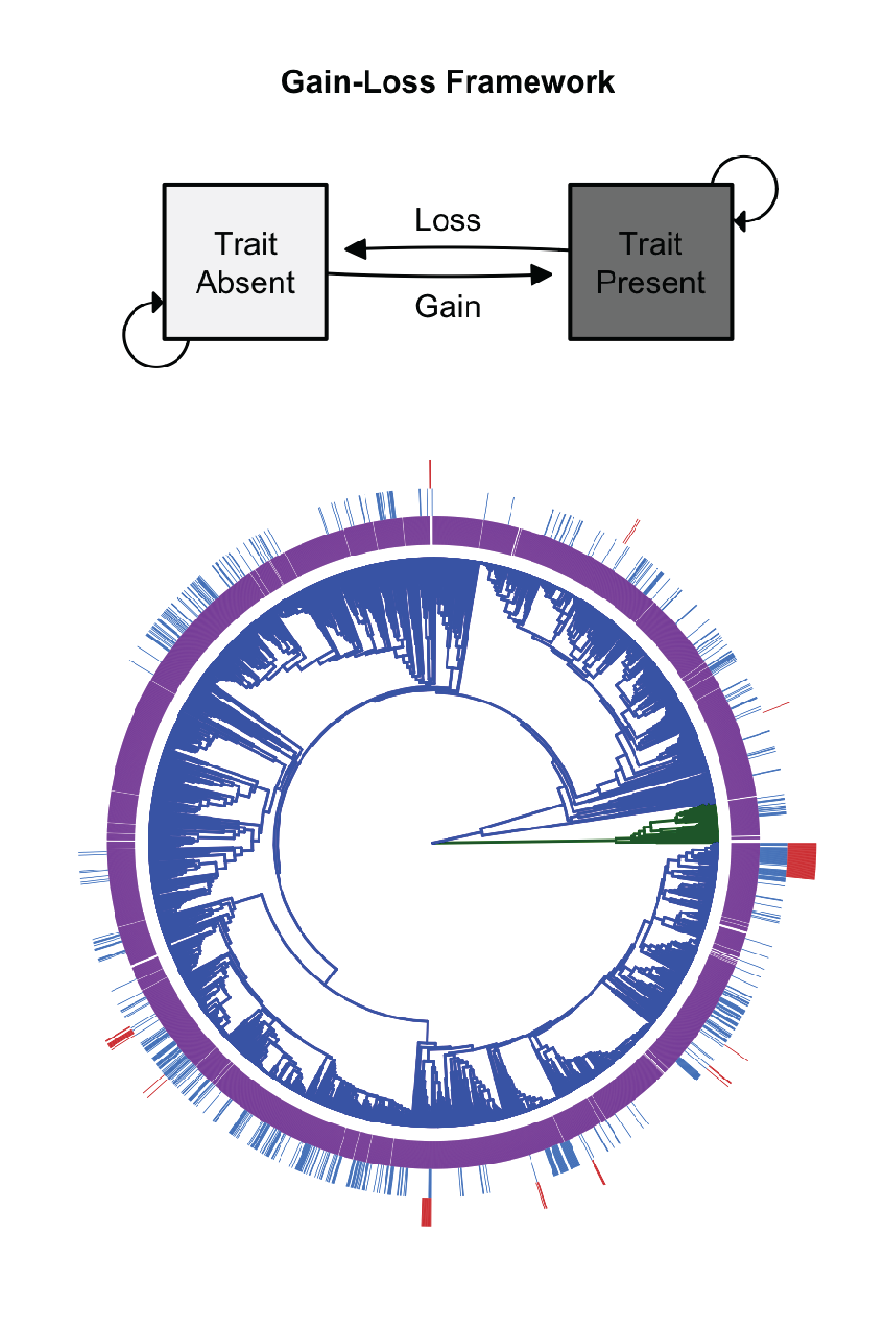
Bacteria and Archaea represent the base of the evolutionary tree of life and contain the vast majority of phylogenetic and functional diversity. These microorganisms and their traits directly impact ecosystems and human health. Using genomic data, we are reconstructing the evolutionary history of microbial traits. In addition, we are determining how gene transition rates can be used to make predictions about the size and type of genes in a genome: generalist genomes comprise many labile genes while specialist genomes comprise more highly conserved functional genes. Our results provide a framework for understanding the evolutionary history of extant microorganisms, and provide insights into the evolution, selective advantage, and phylogenetic patterns of microbial traits.
In collaboration with Dr. Kevin Webster, we are currently expanding this project to better understand the evolutionary history of methanotrophy. We are currently linking the gene and pathway phylogeny to Earth’s geological history to better understand this important ecosystem process.
Related Manuscripts:
- Muscarella ME, O’Dwyer JP (In Revision) Ecological insights from the evolutionary history of microbial innovations. bioRxiv preprint available, DOI: https://doi.org/10.1101/220939
Other Projects
-
Relic DNA
-
Microbial Communities in Evolution Canyon
-
Sampling in Microbial Ecology

